RStudio Conference 2020 Packages and Highlights and Random Packages and thoughts
Sortable
This was part of the poster session in the rstudio::conf() 2020. Great for drag and drop shiny objects from one bucket to other and capture the interaction in the server side.
End-to-End Data Science with Shiny, Plumber and Pins
This was a presentation by Alex Gold (@alexgold) engineer from RStudio. The workflow makes it easier for dploymment and sharing models and input data. This is another way to make the entire process very reproduceable. The files for this presentation is located in github.
Noteworthy package here is pins:

The package library can be found in: pins github directory
Setting up boards are easy in the git hub case you can use:
You can pin the data to your github board using the pin command as follows:
pin(iris, description = "The iris data set", board = "github")You can get the data back using:
pin_get("iris", board = "github")## # A tibble: 150 x 5
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## <dbl> <dbl> <dbl> <dbl> <fct>
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## # … with 140 more rowsOr find the data in the board:
pin_find("iris", board = "github")## # A tibble: 1 x 4
## name description type board
## <chr> <chr> <chr> <chr>
## 1 iris The iris data set table github**The rule is if the data is large (25M in github) and greater then 100M using rsconnect setup avoid using this.
Plumber Workflow

Very well done presentation on how to develop and deploy plumber API with unit tests. Interesting use case for continues deployment from github directly to RStudio Connect. The code base is located under: github and slide decks for the talk are under: Slides
Producing Reproducable Examples

This package is used to generate reproducable examples for stackoverflow or github. Reprex R package is located under: reprex
require(reprex)## Loading required package: reprex(y <- 1:4)## [1] 1 2 3 4mean(y)## [1] 2.5reprex()## No input provided and clipboard is not available.## Rendering reprex...File System
fs package is cross platform file system access package. More user friendly version of the base R. The package location is: fs
rmarkdown series of talks
The first discussion by Atlas the full discussion in located under ratlas. The most interesting aspect of the talk was branded microsoft word branded documents. Interesting no discussion on google branded products from rmarkdown this could be idea for project rgooglemarkdown???
Debugging Keynote
Really interesting talk by Jenney Bryan from RStudio entertaining but useful tips full presentation is under: debugging
ggplot2 Discussions
Great presentation on ggplot2 Slides but same information can be found in the tidyverse documentation Link
The focus of the presentation was around using ggplot within a function and package where input parameters need to become dynamics for generic functions.
Use .data[[“variable_name”]] within aes() and/or vars()
ggplot(…) + item1 + item2 is identical to ggplot(…) + list(item1, item2) Use it to conditinally more than one item to a plot!
Example of passing the col name
require(ggplot2)
col_summary <- function(df, col) {
ggplot(df) +
geom_bar(aes(x = {{ col }})) +
coord_flip()
}
col_summary(mpg, drv)Using the vars within the facet wrap to make it more dynamic
#' @importFrom ggplot2 ggplot aes vars facet_wrap geom_point labs
#'
plot_mpg <- function(colour_var, facet_var) {
mapping <- aes(displ, hwy, colour = .data[[colour_var]])
if (is.null(colour_var)) {
mapping$colour <- NULL
}
if (is.null(facet_var)) {
facet <- NULL
} else {
facet <- facet_wrap(vars(.data[[facet_var]]))
}
ggplot(ggplot2::mpg) +
geom_point(mapping) +
facet
}tidyr

Not directly from the conference but there were bunch of discussions on new tidyr. Since I have been using tidyr the old way trying to explore the new functions. This is mainly using the Hadleys Sept 13 blog post
- pivot_longer() and pivot_wider()
require(tidyr)## Loading required package: tidyriris2<-iris %>% pivot_longer(
-dplyr::starts_with("Spe"),
names_to = c("Type"),
names_pattern = "(.*)"
)
iris2## # A tibble: 600 x 3
## Species Type value
## <fct> <chr> <dbl>
## 1 setosa Sepal.Length 5.1
## 2 setosa Sepal.Width 3.5
## 3 setosa Petal.Length 1.4
## 4 setosa Petal.Width 0.2
## 5 setosa Sepal.Length 4.9
## 6 setosa Sepal.Width 3
## 7 setosa Petal.Length 1.4
## 8 setosa Petal.Width 0.2
## 9 setosa Sepal.Length 4.7
## 10 setosa Sepal.Width 3.2
## # … with 590 more rowstmp<-iris2 %>% dplyr::mutate(Species=as.character(Species),
val=as.numeric(value)) %>%
dplyr::select(-value) %>%
pivot_wider(
names_from = c("Species"),
values_from = val
)
tmp## # A tibble: 4 x 4
## Type setosa versicolor virginica
## <chr> <list> <list> <list>
## 1 Sepal.Length <dbl [50]> <dbl [50]> <dbl [50]>
## 2 Sepal.Width <dbl [50]> <dbl [50]> <dbl [50]>
## 3 Petal.Length <dbl [50]> <dbl [50]> <dbl [50]>
## 4 Petal.Width <dbl [50]> <dbl [50]> <dbl [50]>tmp %>% unnest(everything())## # A tibble: 200 x 4
## Type setosa versicolor virginica
## <chr> <dbl> <dbl> <dbl>
## 1 Sepal.Length 5.1 7 6.3
## 2 Sepal.Length 4.9 6.4 5.8
## 3 Sepal.Length 4.7 6.9 7.1
## 4 Sepal.Length 4.6 5.5 6.3
## 5 Sepal.Length 5 6.5 6.5
## 6 Sepal.Length 5.4 5.7 7.6
## 7 Sepal.Length 4.6 6.3 4.9
## 8 Sepal.Length 5 4.9 7.3
## 9 Sepal.Length 4.4 6.6 6.7
## 10 Sepal.Length 4.9 5.2 7.2
## # … with 190 more rows- unnest_auto(), unnest_longer(), unnest_wider(), and hoist()
tmp %>% unnest_longer(setosa)## # A tibble: 200 x 4
## Type setosa versicolor virginica
## <chr> <dbl> <list> <list>
## 1 Sepal.Length 5.1 <dbl [50]> <dbl [50]>
## 2 Sepal.Length 4.9 <dbl [50]> <dbl [50]>
## 3 Sepal.Length 4.7 <dbl [50]> <dbl [50]>
## 4 Sepal.Length 4.6 <dbl [50]> <dbl [50]>
## 5 Sepal.Length 5 <dbl [50]> <dbl [50]>
## 6 Sepal.Length 5.4 <dbl [50]> <dbl [50]>
## 7 Sepal.Length 4.6 <dbl [50]> <dbl [50]>
## 8 Sepal.Length 5 <dbl [50]> <dbl [50]>
## 9 Sepal.Length 4.4 <dbl [50]> <dbl [50]>
## 10 Sepal.Length 4.9 <dbl [50]> <dbl [50]>
## # … with 190 more rowstmp %>% unnest_wider(setosa)## # A tibble: 4 x 53
## Type ...1 ...2 ...3 ...4 ...5 ...6 ...7 ...8 ...9 ...10 ...11
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Sepal.… 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 5.4
## 2 Sepal.… 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 3.7
## 3 Petal.… 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 1.5
## 4 Petal.… 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 0.2
## # … with 41 more variables: ...12 <dbl>, ...13 <dbl>, ...14 <dbl>,
## # ...15 <dbl>, ...16 <dbl>, ...17 <dbl>, ...18 <dbl>, ...19 <dbl>,
## # ...20 <dbl>, ...21 <dbl>, ...22 <dbl>, ...23 <dbl>, ...24 <dbl>,
## # ...25 <dbl>, ...26 <dbl>, ...27 <dbl>, ...28 <dbl>, ...29 <dbl>,
## # ...30 <dbl>, ...31 <dbl>, ...32 <dbl>, ...33 <dbl>, ...34 <dbl>,
## # ...35 <dbl>, ...36 <dbl>, ...37 <dbl>, ...38 <dbl>, ...39 <dbl>,
## # ...40 <dbl>, ...41 <dbl>, ...42 <dbl>, ...43 <dbl>, ...44 <dbl>,
## # ...45 <dbl>, ...46 <dbl>, ...47 <dbl>, ...48 <dbl>, ...49 <dbl>,
## # ...50 <dbl>, versicolor <list>, virginica <list>tmp %>% unnest_auto(setosa)## # A tibble: 200 x 4
## Type setosa versicolor virginica
## <chr> <dbl> <list> <list>
## 1 Sepal.Length 5.1 <dbl [50]> <dbl [50]>
## 2 Sepal.Length 4.9 <dbl [50]> <dbl [50]>
## 3 Sepal.Length 4.7 <dbl [50]> <dbl [50]>
## 4 Sepal.Length 4.6 <dbl [50]> <dbl [50]>
## 5 Sepal.Length 5 <dbl [50]> <dbl [50]>
## 6 Sepal.Length 5.4 <dbl [50]> <dbl [50]>
## 7 Sepal.Length 4.6 <dbl [50]> <dbl [50]>
## 8 Sepal.Length 5 <dbl [50]> <dbl [50]>
## 9 Sepal.Length 4.4 <dbl [50]> <dbl [50]>
## 10 Sepal.Length 4.9 <dbl [50]> <dbl [50]>
## # … with 190 more rows- nest() and unnest() moved to (pack()/unpack(), and chop()/unchop())
df <- tibble(x1 = 1:3, x2 = 4:6, x3 = 7:9, y = 1:3)
df## # A tibble: 3 x 4
## x1 x2 x3 y
## <int> <int> <int> <int>
## 1 1 4 7 1
## 2 2 5 8 2
## 3 3 6 9 3df %>% pack(x = starts_with("x")) %>% unpack(x)## # A tibble: 3 x 4
## y x1 x2 x3
## <int> <int> <int> <int>
## 1 1 1 4 7
## 2 2 2 5 8
## 3 3 3 6 9- new expand_grid()
students <- tribble(
~ school, ~ student,
"A", "John",
"A", "Mary",
"A", "Susan",
"B", "John"
)
expand_grid(students, semester = 1:2)## # A tibble: 8 x 3
## school student semester
## <chr> <chr> <int>
## 1 A John 1
## 2 A John 2
## 3 A Mary 1
## 4 A Mary 2
## 5 A Susan 1
## 6 A Susan 2
## 7 B John 1
## 8 B John 2slider
Great new package not yet in cran but simplifies and replaces some of the tibbletime functionality. Great for applications such as rolling sd and rolling correlation
Good write-up in the slider website simple example below:
library(slider)
library(tidyverse)## ── Attaching packages ────────────────────────── tidyverse 1.2.1 ──## ✓ ggplot2 3.2.1 ✓ purrr 0.3.2
## ✓ tibble 2.1.3 ✓ dplyr 0.8.3
## ✓ readr 1.3.1 ✓ stringr 1.4.0
## ✓ ggplot2 3.2.1 ✓ forcats 0.3.0## ── Conflicts ───────────────────────────── tidyverse_conflicts() ──
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()set.seed(123)
df <- tibble(
y = rnorm(100),
x = rnorm(100)
)
out<-df %>% mutate(m=slide2_dbl(y,x,cor, .before = 19,.complete = T))
tail(out)## # A tibble: 6 x 3
## y x m
## <dbl> <dbl> <dbl>
## 1 1.36 -1.31 -0.141
## 2 -0.600 2.00 -0.204
## 3 2.19 0.601 -0.0799
## 4 1.53 -1.25 -0.154
## 5 -0.236 -0.611 -0.114
## 6 -1.03 -1.19 -0.00903usethis
There was a lot of discussions on usethis package for package building setup and deployment. The link to usethis package is here: usethis
There is also a good blogpost on the steps to setup a R package quickly: Package blogpost
testthat
This was another workflow package which forces good software engineering. This will work in concert with usethis and other package building tools. Good write up by Hadley and package library for testthat
scales

Scales
Presentation on the functionality of scales from scale. The slides for this presentation highlight 5 areas of functionality for scales package.
- transformation
- bounds and rescaling
- breaks
- labels
- palettes
A good usecase is to create custom transformation:
require(scales)## Loading required package: scales##
## Attaching package: 'scales'## The following object is masked from 'package:purrr':
##
## discard## The following object is masked from 'package:readr':
##
## col_factorrequire(ggplot2)
# use trans_new to build a new transformation
dollar_log <- trans_new(
name = "dollar_log",
# extract a single element from another trans
trans = log10_trans()$trans,
# or write your own custom functions
inverse = function(x) 10^(x),
breaks = breaks_log(),
format = label_dollar()
)ggplot(diamonds, aes(y = price, x = carat)) +
geom_hex() +
scale_y_continuous(trans = dollar_log) +
scale_x_log10()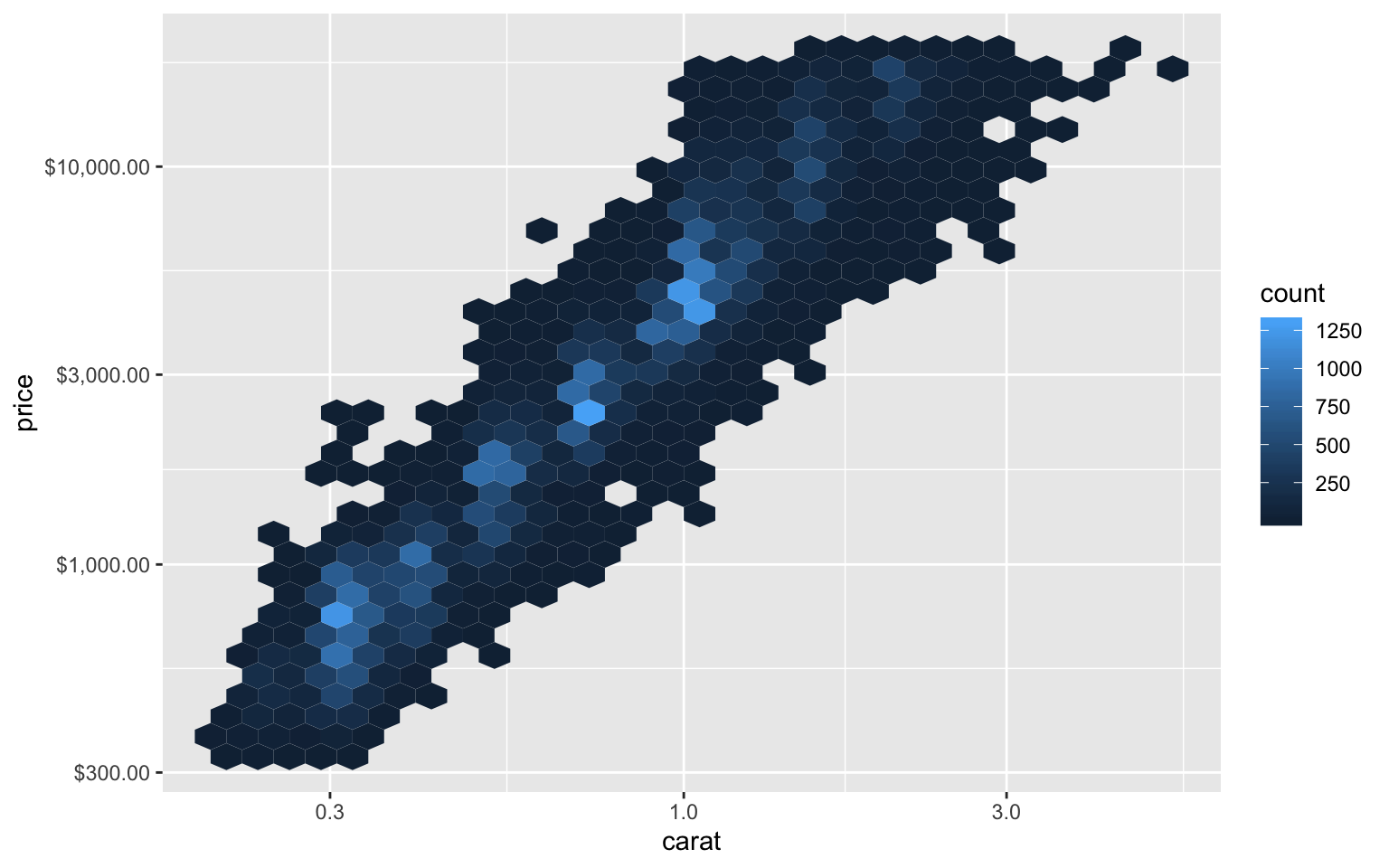
or rescaling data:
squish(c(-1, 0.5, 1, 2, NA), range = c(0, 1))## [1] 0.0 0.5 1.0 1.0 NAdiscard(c(-1, 0.5, 1, 2, NA), range = c(0, 1))## [1] 0.5 1.0 NAcensor(c(-1, 0.5, 1, 2, NA), range = c(0, 1))## [1] NA 0.5 1.0 NA NAggplot(iris, aes(x = Sepal.Length, y = Sepal.Width,
colour = Sepal.Length)) +
geom_point() +
scale_color_continuous(limit = c(6, 8), oob = scales::squish)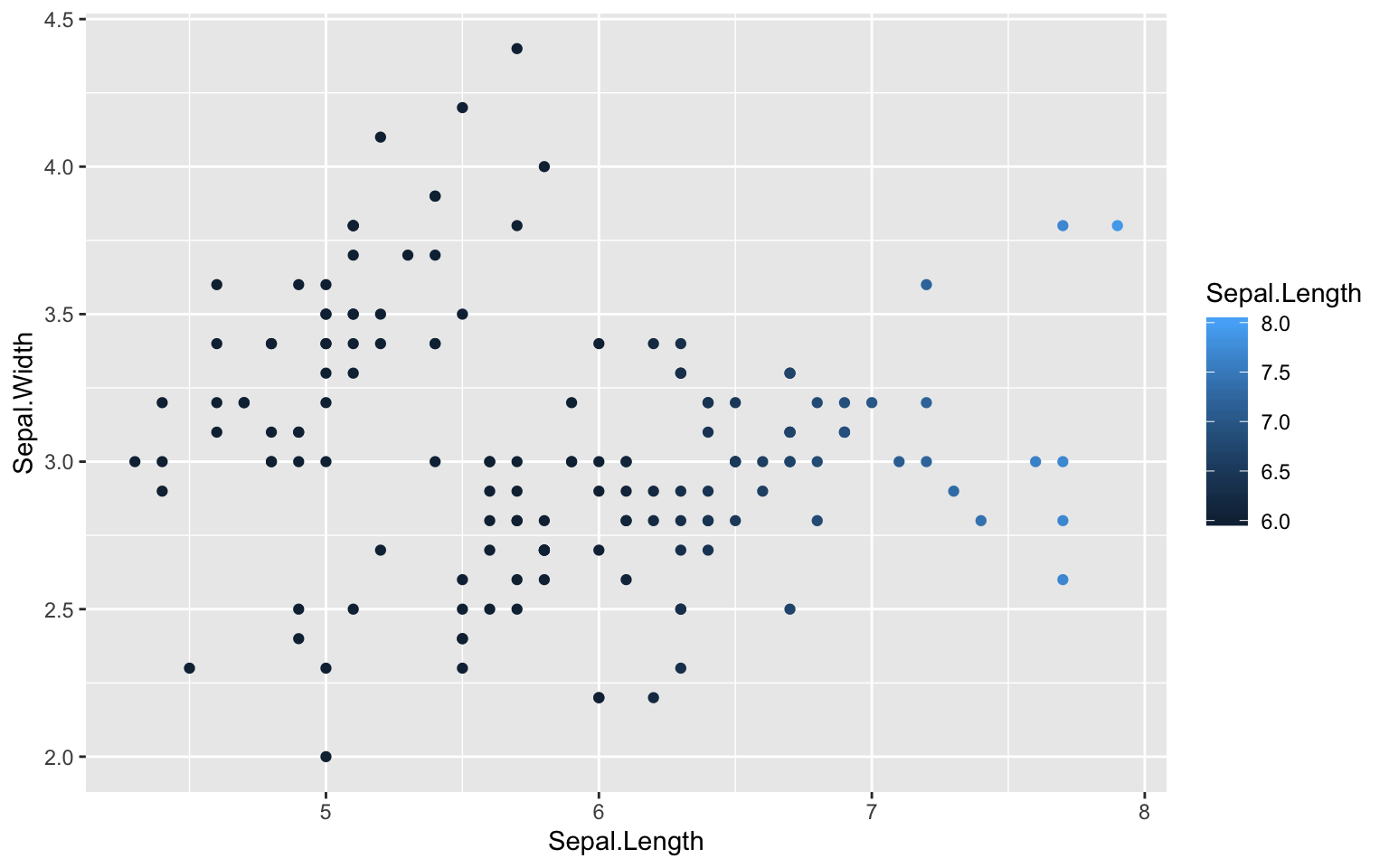
tidyeval
Using tidyeval framework for metaprogramming in R. It is commonly used in tidyverse to apply data masking. There is good writeup and bookdown is uder tidyevaluation.
The new innovation here is tunnelling using brakets’
require(tidyverse)
mean_by1<-function(data,by,var) {
data %>%
group_by(by) %>%
summarize(var:= mean(var))
}
#mean_by1(iris,Species,Petal.Width)
mean_by<-function(data,by,var) {
data %>%
group_by({{ by}}) %>%
summarize({{var}} := mean({{var}}))
}
mean_by(iris,Species,Petal.Width)## # A tibble: 3 x 2
## Species Petal.Width
## <fct> <dbl>
## 1 setosa 0.246
## 2 versicolor 1.33
## 3 virginica 2.03Time series forecasting workshop
Next week blog post.
This is for the next blogpost the github for the training sessions github
Text book is under timeseries forecasting
renv
New blog post idea: renv
tidymodels/tune
New blog post idea: tidymodels/tune
tfprobability
New blogpost idea: tfprobability